Classification#
Classification is the process of grouping images into predetermined categories. Classification can be used to discriminate images based on user preference, e.g. different cell types, phenotypes or tissue types etc.
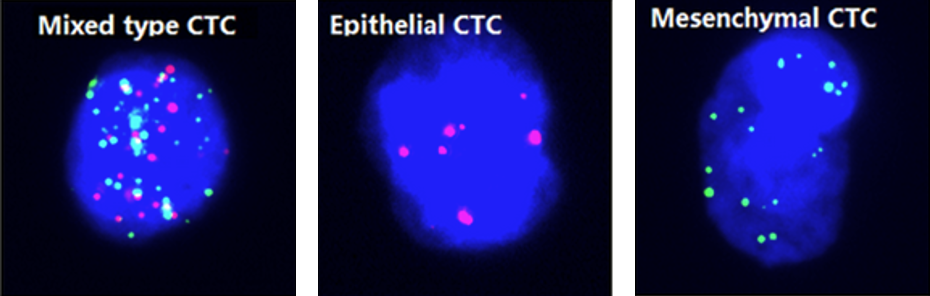
Classification of different types of circulating tumor cells (CTCs) [1]#
Labelling#
To train a neural network for classification it is necessary to create images that are labelled accordingly.

Object detection tools#
Different tools and functions in MIA exist to label images for classification.
To label an image select a tool and draw inside the image to label image objects.
Note
There is no save option as all changes during labelling are saved immediately. Use  or ctrl + z to undo the last labelling action.
or ctrl + z to undo the last labelling action.
All labels are saved in a subfolder inside the currently active folder and have the same file name as the currently selected image, but with .npz as file extension.
Tools#
As classification only needs a single label for each image, the tools are simpler compared to other applications and you do not have to explicitly click inside the image.
By pressing  or F1 currently selected class is assigned to the image.
or F1 currently selected class is assigned to the image.
By pressing  or F2 currently selected class is assigned to the image and the next image is selected.
or F2 currently selected class is assigned to the image and the next image is selected.
Tip
The classification label is shown as a border around the image with the color of the corresponding class
Training#
For details about neural network training see Training.
Neural Network architectures#
For classification there is no extra architecture implemented, meaning that the architecture is identical to the backbone.
Tip
All architectures are implemented without fully connected layers, even if the original architecture had fully connected layers (like vgg-nets).
The following backbones are currently supported, untrained or with pretrained weights pre-trained on imagenet dataset [2]:
Model |
Options |
Ref. |
DenseNet |
densenet121, densenet169, densenet201 |
|
EfficientNet |
efficientnetb0, efficientnetb1, efficientnetb2, efficientnetb3, efficientnetb4, efficientnetb5, efficientnetb6, efficientnetb7 |
|
Inception |
inceptionv3, inceptionresnetv2 |
|
MobileNet |
mobilenet, mobilenetv2 |
|
NasNet |
nasnetlarge, nasnetmobile |
|
ResNet |
resnet18, resnet34, resnet50, resnet101, resnet152, resnet50v2, resnet101v2, resnet152v2 |
|
ResNeXt |
resnext50, resnext101 |
|
SE-ResNet |
seresnet18, seresnet34, seresnet50, seresnet101, seresnet152 |
|
SE-ResNeXt |
seresnext50, seresnext101 |
|
SENet |
senet154 |
|
VGG |
vgg16, vgg19 |
|
Xception |
xception |
Tip
Generally the numbers behind the backbone architecture gives either the number of convolutional layers (e.g. resnet18) or the model version (e.g. inceptionv3).
When you have limited computing recources use a small network architecture or a network optimized for efficiency (e.g. mobilenetv2).
From the supported network-backbones the nasnetlarge shows the highest performance on imagenet classification.
From the supported network-backbones the mobilenet has the fastest processing time and fewest parameters.
Losses and Metrics#
For classification several objective function have been tested for neural network optimization and directly impact the model training.
Metrics are used to measure the performance of the trained model, but are independent of the optimization and the training process.
The loss and metric functions can be set in  Train Model →
Train Model →  Settings.
Settings.
Cross Entropy#
The cross entropy loss is a widely used objective function used for classification. It is defined as:
with \(p_i\) the true label and \(q_i\) the model prediction for the \(i_{th}\) class.
Focal Loss#
The focal loss is an extension of the cross entropy, which improves performance for unbalanced datasets [13]. It is defined as follows:
with \(\gamma\) as the focussing parameter. Default is set \(\gamma = 2\).
Kullback-Leibler Divergence#
The Kullback-Leibler Divergence, sometimes referred as relative entropy, is defined as follows:
Accuracy#
The pixel accuracy measures all images that are classified correctly:
with \(t_p\) the true positives (\(p_i=1\) and \(q_i=1\)), \(t_n\) the true negatives (\(p_i=0\) and \(q_i=0\)), \(f_p\) the false positives (\(p_i=0\) and \(q_i=1\)) and \(f_n\) the false negatives (\(p_i=1\) and \(q_i=0\)). The accuracy is a misleading measure for imbalanced data.
Precision#
The precision measures how many of all predicted positives, actually are positve:
Recall#
The recall measures how many of the actual positives, are predicted as positve:
F1-Score#
The F1-score is a balanced measure of precision and recall